I am a wildlife geneticist that utilizes genomics, GIS technology, and a variety of informatic techniques to examine how environmental heterogeneity impacts genomic variation in wildlife. My work covers many species and questions due to the varied impacts of environmental heterogeneity, from how past climatic events influence biodiversity to the conservation genetics of imperiled species and population dynamics of overabundant mammals. Being at the museum, I am also very passionate about incorporating museum collections into my research.

Cryptic Biodiversity
Range-wide Phylogeography
My work in phylogeography has largely focused on widespread mammals like rodents, carnivores, and ungulates. Despite these species’ tendencies to be generalists and highly mobile, evolutionary histories vary widely. For example, I have documented deep divergence between lineages in gray fox and little structure in American badgers (left). Identifying areas of secondary contact and hybridization between these lineages are also important for understanding reproductive isolation and adaptation of evolutionary lineages, so much of my phylogenetic work also focuses on hybrid zones in North America (e.g., Great Plains and Pacific Northwest). Going forward, genomics offers an exciting toolkit to investigate adaptive variation and gene flow within evolutionary lineages, and ultimately examine evolutionary mechanisms across time and space.
Conservation Genetics
Examining human impacts on imperiled populations
Losses in connectivity and genetic diversity are major concerns for imperiled species. Both connectivity and genetic diversity are considered essential for healthy populations, so my work uses genetic data to evaluate potential threats to conservation concern species. I previously focused on identifying landscape factors that impact connectivity, but more recently, my conservation work involves estimating population densities, inbreeding, and parentage in endangered species like mountain gorillas and black rhinoceros. More locally, I often collaborate with management agencies to assess conservation status of conservation concern species like Allegheny woodrats and Buxton woods white-footed mice.
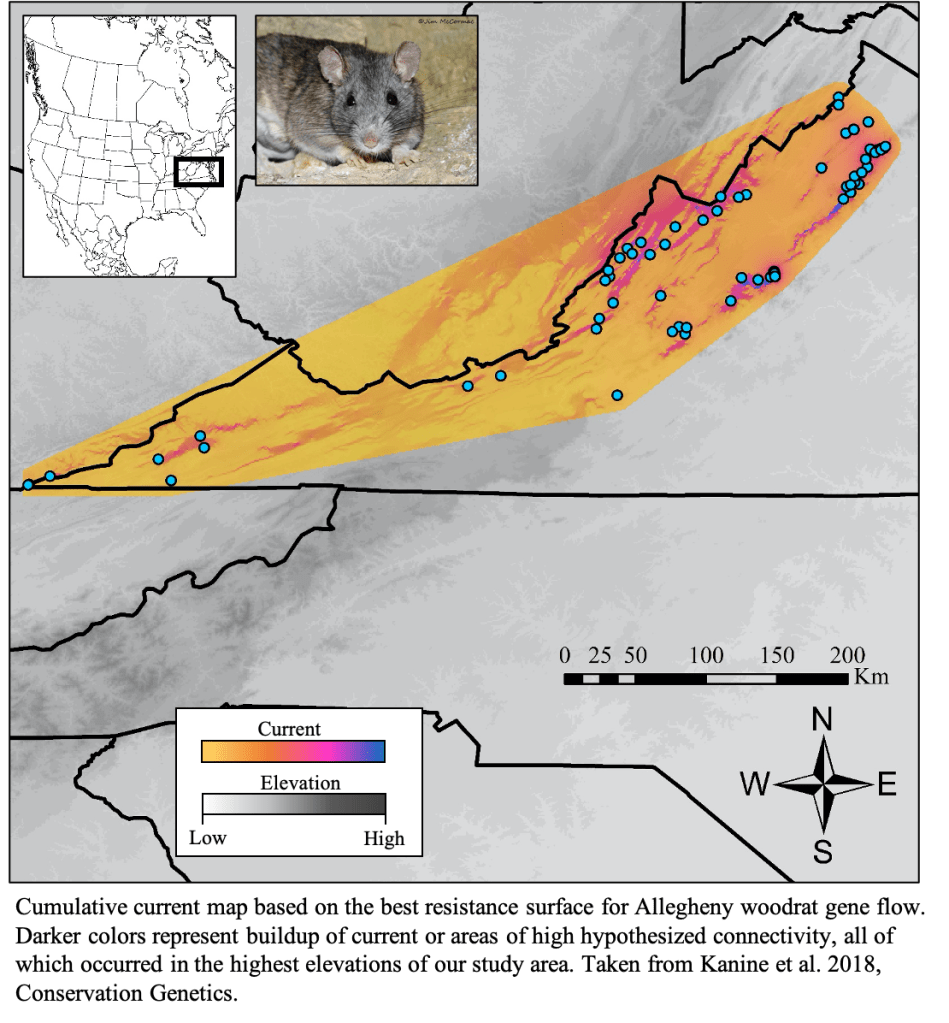

Population monitoring
Non-invasive estimation via fecal DNA
Another aspect of my research is monitoring of abundant species that benefit from humans. Invasive and overly abundant species, for example, often were either introduced by humans or utilize modified landscapes. These species can be a major threat to biodiversity by direct competition or predation on native ecosystems and increase human-wildlife conflict. Thus, my work seeks to evaluate management actions that aim to control overly abundant (e.g., coyotes on the Outer Banks, North Carolina and white-tailed deer in Durham County).
Rapid Adaptation
Genomic change in the Anthropocene
Global change is rapidly changing native ecosystems through processes like urbanization, altered climate, pollution, and even disease. Species must rapidly adapt to these changes or face extinction. My current work uses high resolution whole genome data to examine the effects of urbanization and rabies on raccoons and striped skunks across North America. Both species thrive in urban environments and serve as a reservoir for unique strains of rabies virus, but little is known about potential genomic mechanisms that may underpin their adaptation to these strong selective pressures.

Selected Publications
Kierepka EM, Preckler-Quisquater S, Reding DM, Piaggio AJ, Riley SPD, Sacks BN (2023)
Genomic analyses of gray fox lineages suggest ancient divergence and secondary contact in the southern Great Plains. Journal of Heredity 114: 110-119.
Sacks BN, Hu T, Kierepka EM, Vanderzwan SL, Hickey JL (2021) Development of 79 SNP markers to individually genotype and sex-type endangered mountain gorillas (Gorilla beringei beringei). Conservation Genetics Resources 13: 375-377.
Kierepka EM, Anderson SJ, Swihart RK, Rhodes Jr OE (2020) Differing landscape effects on genetic diversity and differentiation in eastern chipmunks. Heredity 124: 457-468.
Davis AJ, Keiter DA, Kierepka EM, Slootmaker C, Piaggio AJ, Beasley JC, Pepin KM (2020) A comparison of cost and quality of three methods for estimating density for wild pig (Sus scrofa). Scientific Reports 10: 1-14.
Kierepka EM, Juarez R, Turner K, Smith J, Hamilton M, Lyons P, Hall MA, Beasley JC, Rhodes Jr OE (2019) Population genetics of invasive brown tree snakes (Boiga irregularis) on Guam, USA. Herpetologica 75: 208-217.
Kierepka EM, Kilgo JC, Rhodes Jr OE (2017) Effect of compensatory immigration on the genetic structure of coyotes. The Journal of Wildlife Management 81: 1394-1407.
Kierepka EM, Latch EK (2016) High gene flow in the American badger overrides habitat preferences and limits broadscale genetic structure. Molecular Ecology 25: 6055-6076.